data.table

What is data.table and why use it?
- It’s the high performance version of base R’s data.frame for manipulation
- Very fast
- Memory efficient
- Lots of features similar to dplyr and other tidyverse functions
- Great support and documentation + no other dependencies
- Downsides?
- Can be complex to learn
- Less intuitive than the tidyverse
- Less niche features compared to the tidyverse (ie. The complete() function)
- Isn’t a complete ‘verse’ – ie. No plotting
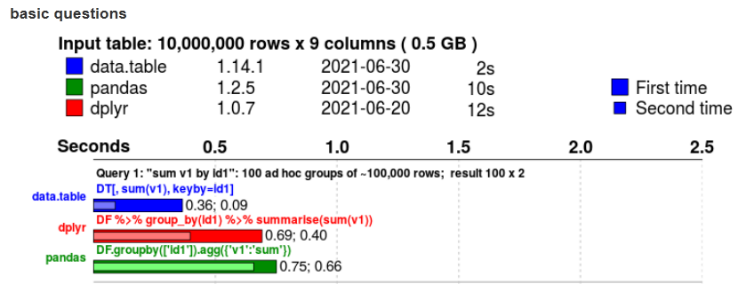
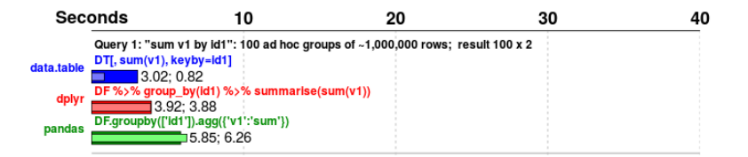
How fast is it?
500MB
5GB
50GB

Not only faster but less onboard memory used https://h2oai.github.io/db-benchmark/
What can it do?
Most of the functions of dplyr and a bit more:
Manipulate
Filter
Sort
Compute
Group by
Bind rows / columns
Joining columns
Wide – Long format
Read + Write files
Almost everything you’d need to reshape data
Loading in and writing out data

- Base/ Tidyverse R: read.csv() or read.xls() -> Becomes a data frame
- Works well and has a viewer for easy column determination
- Can change to a data.table by setDT () or as.data.table ()
- Similar write.csv() etc. to write it to a file.

- data.table: fread () -> Straight into a data.table format
- Functions identically to a data.frame
- Very fast to read in data and doesn’t load it all onto RAM (lazy loading)
- Can read zipped and compressed files without saving a decompressed file
- Use fwrite () to also write out data much faster (I’ve come across issues here)
If this is the only function you want, use “vroom” instead
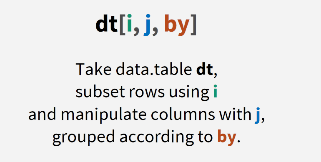
Syntax (The hard part!)

tidyverse
Ethos: Do one thing at a time
Each function has an easy to understand name
Example:
The basic data.table syntax
Example dataset:
| Fruit | Variety | Weight |
|---|---|---|
| Orange | Navel | 400 |
| Apple | Jazz | 300 |
| Apple | Jazz | 400 |
| … | … | … |
Filter a row:
Example[Fruit == "Orange"]
Filter and add a row:
Example[Fruit == "Apple", Fruit_Variety := paste(Variety,Fruit)]
Just add a row:
Example[,BaseCost := (Weight/1000) * 2]
Perform a grouped summary
Example[, by = "Fruit", Fruit_Weight := mean(Weight)]
Moving from the tidyverse to data.table
- Calling a data.table or data.frame: The same (almost)
- In the tidyverse you can call the df within the function ie. select( df _ , …) _ or before df _ %>% select()_ . In data.table it’s only df[…]
- Manipulating data:
- In the tidyverse you just use = , ie. mutate( df , ColB _ = _ ColA _ * 10)_
- In data.table you use := or .() , ie. df [,.( ColB _ = _ ColA _ * 10 )] _ or _ _ df [,:= …]
- Important to note above df [, … ] not df […] because this tries to subset (filter)
- Chaining functions:
- In the tidyverse you use the pipe %>%
- In data.table you can use %>% but better to use dt specific chaining df […][…][…]
- Using %>% looks like df […] %>% .[…] %>% .[…]
Chaining in data.table
Why Chain? Lots of concise alterations, and less objects saved
- data.table specific chaining looks like df […][…][…]
“I want to know all the types of fruit that will cost on average above $1 per item in order of most to least expensive”
Example2 <- Example[, Fruit_Weight := mean(Weight), by = "Fruit"][, ItemCost := (Weight/1000) * Cost.kg][ItemCost >= 1]\[order(Item)]
- Note: mutate( df , A = 1, B = A + 2) is not the same as df [,.(A = 1, B = A + 2)]
Tidyverse
Example2 <- Example %>%
group_by(Fruit) %>%
summarise(Fruit_Weight = mean(Weight)) %>%
Ungroup() %>%
Mutate(ItemCost = (Weight/1000) * Cost.kg) %>%
Filter (ItemCost >= 1) %>%
Arrange(Item)data.table with pipes
Example dataset:
| Fruit | Variety | Weight | Cost.kg |
|---|---|---|---|
| Orange | Navel | 400 | 2 |
| Apple | Jazz | 300 | 4 |
| Apple | Gala | 400 | 4.5 |
| … | … | … |
Some similar functions:

filter() ,mutate(), group_by ()
summarise(df, sum(ColA), sd (ColB))
arrange(df, ColA)
select(df, ColA, ColB, ColD)
group_by(df, ColA)
gather()
spread()
full_join()

Intrinsic df[ filter, mutate, by = ]
df[,.(sum = ColA, sd(ColB))]
df[order(ColA)]
df[,.(ColA, ColB, ColD)]
df[,by = ColA] or df[,keyby = ColA]
melt()
dcast()
merge(all = “true”)
The lazy way – improve your code sooner

You: *“I really want to use _ data.table* _ to speed things up but I don’t have time to learn it or alter my pre-existing code”
Me: Give dtplyr a try?
dtplyr allows you to write dplyr code that is automatically translated to the equivalent data.table code under the hood.
Just load the package library( dtplyr ) and use df2 <- lazy_dt ( df ) before performing your normal operations. At the end use as.data.table () or as_tibble ()

Final notes
Some code doesn’t translate well between the two – Try to check on your data frames often especially when you start using this
Be careful with ordering, grouping, and manipulating or joining.
- The speed of data.table can deceive you into thinking you performed a simple normal mutation instead of creating 100x more rows with duplications everywhere
Running some package functions within data.table that don’t run efficiently won’t be much faster. Ie. The package creator wrote the code using tidyverse dependencies
Extra Resources:
Package information page:
https://rdatatable.gitlab.io/data.table/
Github wiki:
https://github.com/Rdatatable/data.table/wiki/Getting-started
A great comparison website between data.table and dplyr (I use this a lot):